popcycle
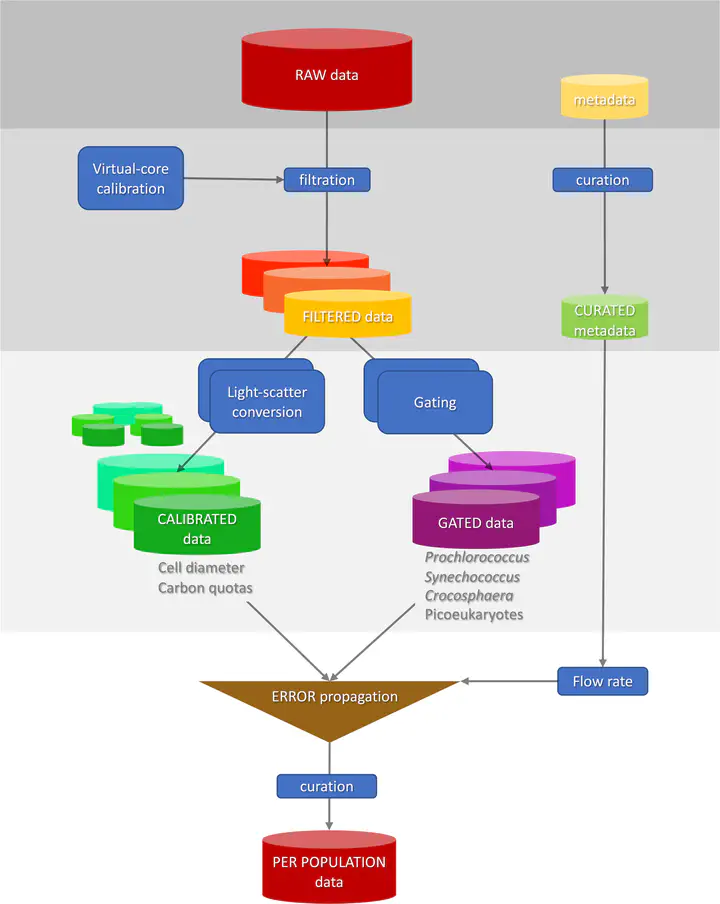
An R package that offers a reproducible approach to process, calibrate and curate SeaFlow data.
Code is available here: https://github.com/seaflow-uw/popcycle
Description of the model is available here: https://doi.org/10.1038/s41597-019-0292-2